
Recently, researchers in the Institute of Bast Fiber Crops, Chinese Academy of Agricultural Sciences (IBFC, CAAS) have reported a comprehensive map of garlic genomic variation, which can indicate the garlic population diverged at least 100,000 years ago, and the two groups cultivated in China were domesticated from two independent routes with differential selections for the bulb traits.This study provides valuable resources for garlic genomics-based breeding, and comprehensive insights into the evolutionary history of this clonal-propagated crop.
Garlic is an entirely sterile crop with important value as a vegetable, condiment, and medicine. However, the evolutionary history of garlic remains largely unknown. Researchersfrom IBFC report a comprehensive map of garlic genomic variation, consisting of amazingly 129.4 million variations. The garlic population diverged at least 100,000 years ago, and the two groups cultivated in China were domesticated from two independent routes. Consequently, 15.0% and 17.5% of genes underwent an expression change in two cultivated groups, causing a reshaping of their transcriptomic architecture.Furthermore, researchers find independent domestication leads to few overlaps of deleterious substitutions in these two groups due to separate accumulation and selection-based removal. By analysis of selective sweeps, genome-wide trait associations and associated transcriptomic analysis,differential selections for the bulb traits in these two garlic groups during their domestication have been uncovered.
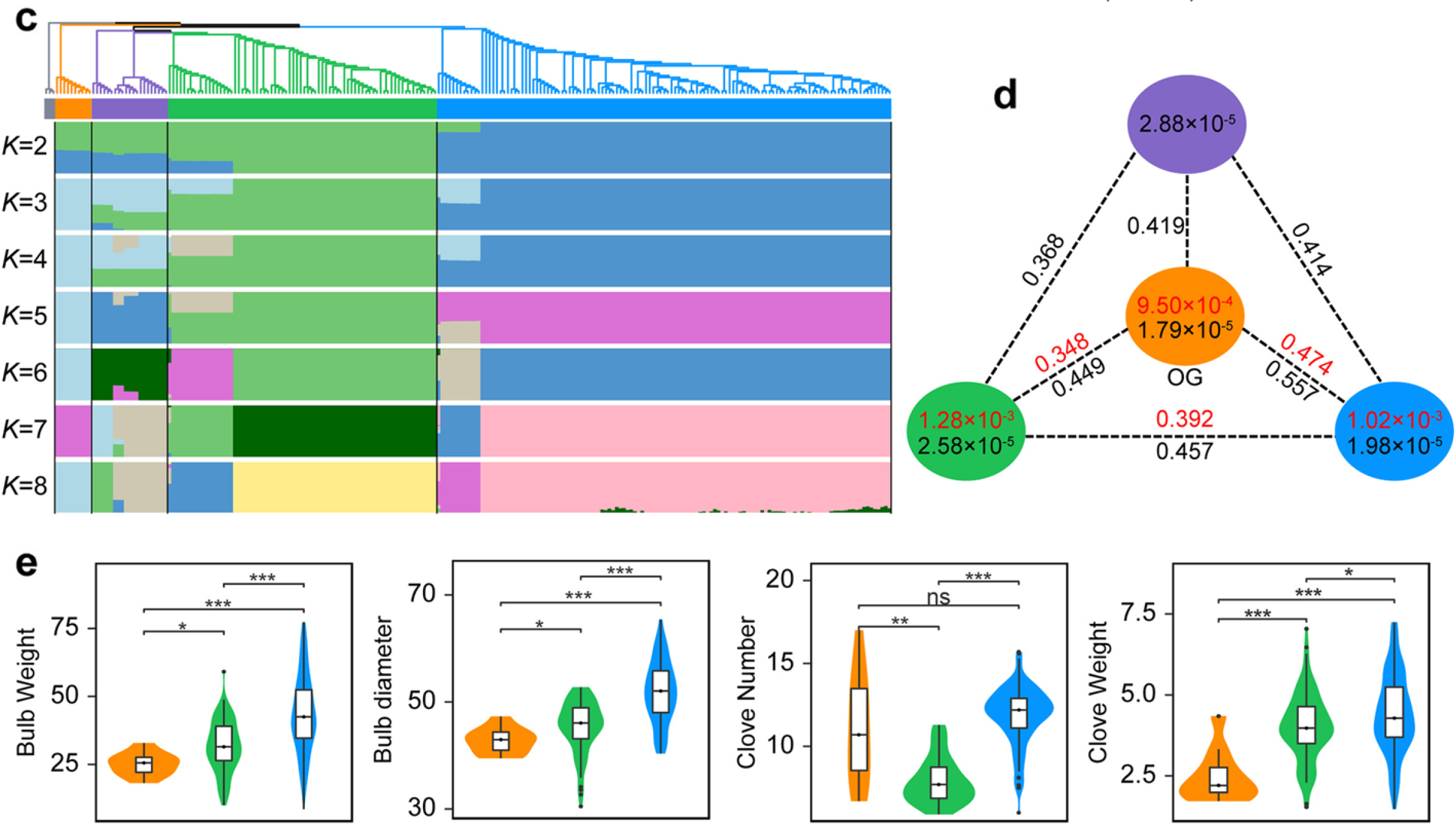
Fig. Population structure and genetic diversity of garlic accessions
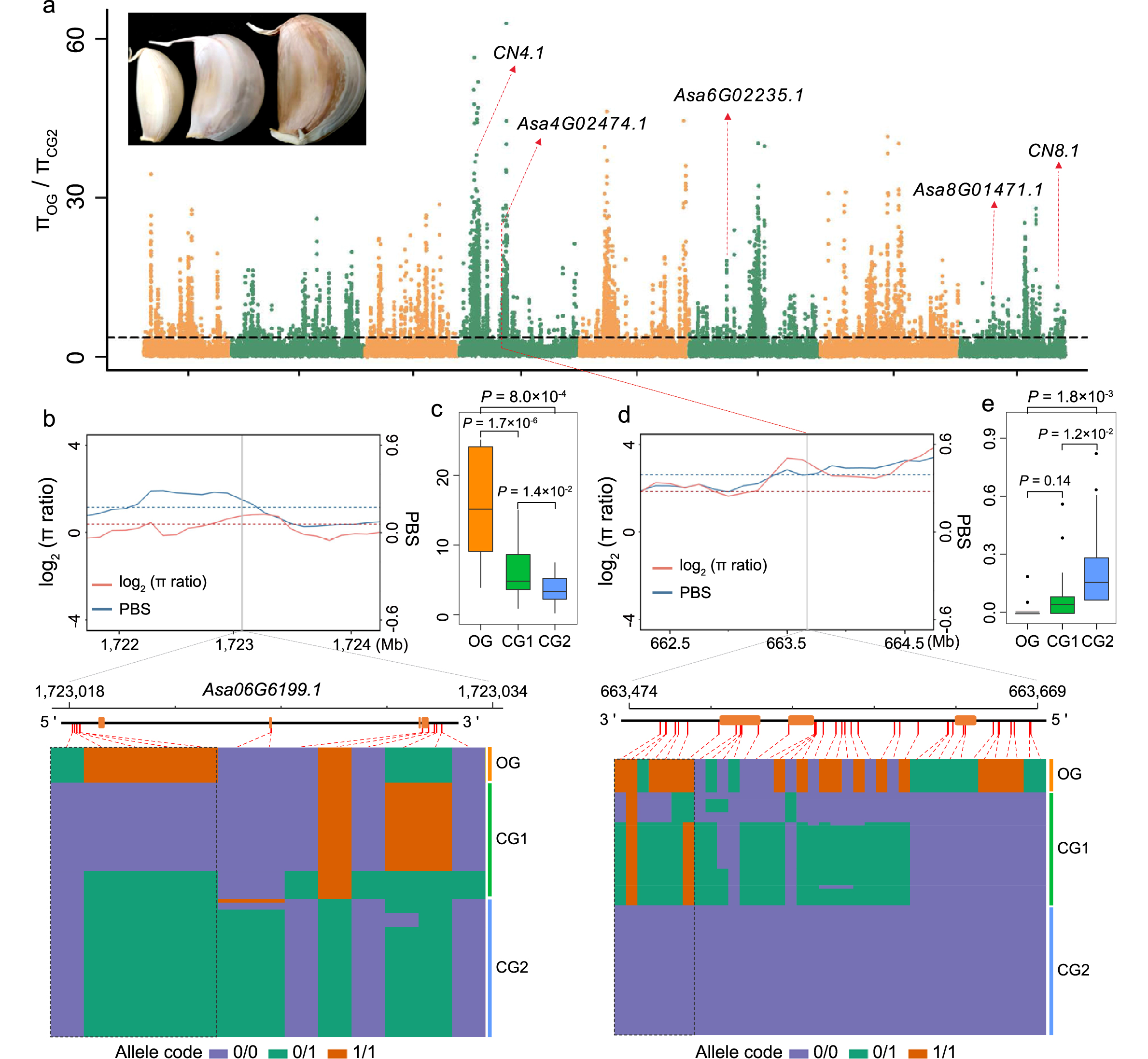
Fig.Selection signatures for the clove weight in CG2
The research was supported by the Science and Technology Innovation Project of the CAAS, the General Program of the National Natural Science Foundation of China and the Shandong Provincial Key Research and Development Program.
The study entitledGenomic insights into the evolutionary history and diversifcation of bulb traits in garlichas been published online in Genome Biology and can be accessed through the following link https://genomebiology.biomedcentral.com/articles/10.1186/s13059-022-02756-1.

